CCS‐Consensuser: A Haplotype‐Aware Consensus Generator for PacBio Amplicon Sequences
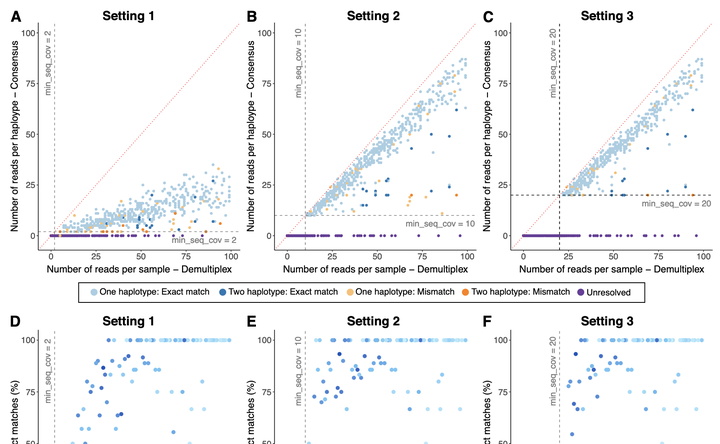 Image credit: C. Congrains
Image credit: C. CongrainsAbstract
DNA sequencing technology has undergone substantial improvements in recent years, to the extent that Third Generation Sequencing platforms are capable of massively generating long-reads. Amplicon sequencing has been among the most popular techniques due to its wide application in diverse fields of biological sciences. However, there is a lack of software specifically designed to analyse intra-individual genetic variation using amplicon long-read data. Here, we present CCSconsensuser, an end-to-end pipeline that generates consensus sequences from amplicon sequencing using high-fidelity reads produced by PacBio circular consensus sequencing (CCS). We evaluated the concordance of the results produced using CCS + CCS-consensuser and other sequencing platforms (Illumina and Sanger), as well as accuracy using a simulated dataset. This assessment showed that CCS amplicon data coupled with CCS-consensuser can produce high-quality sequences (PHRED > 30). The pipeline resulted in high proportions of identical sequence bins for real data, achieving up to 94.94% concordance with COI Sanger sequences and 92.61% with nuclear loci Illumina sequences (considering heterozygous loci), and 95.55% with a fully phased nuclear simulated dataset. Furthermore, our pipeline can be used to detect heteroplasmy in mtDNA, cross-contamination, resolve the phase of nuclear genes in diploid organisms, and conceivably for multi-copy gene systems such as rDNA. These results not only support its potential for application in studies using haploid data such as DNA barcoding, but also demonstrate its unique capacity to explore within individual haplotype variation. Therefore, our strategy shows promise for a broad range of applications in biology and medicine that have been challenging to assess using traditional techniques.