DNA barcodes and reliable molecular identifications in a diverse group of invasive pests: lessons from Bactrocera fruit flies on variation across the COI gene, introgression, and standardization
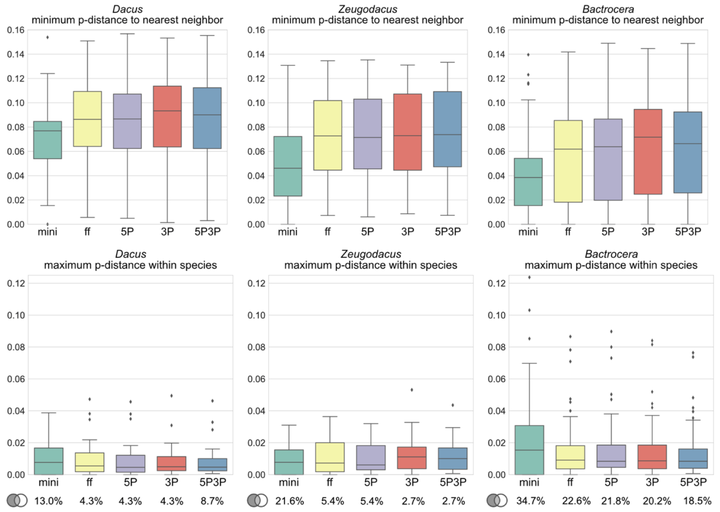 Image credit: C Doorenweerd
Image credit: C DoorenweerdAbstract
The utility of a universal DNA ‘barcode’ fragment of 658 base pairs of the Cytochrome C Oxidase I (COI) gene for the recognition of all animal species has been a widely debated topic on theoretical and practical levels. Regardless of its challenges, large amounts of COI sequence data have been produced in the last two decades. To optimally use the data towards reliable species identification will require further steps to validate the method and reference libraries. The fruit fly tribe Dacini holds about a thousand species, of which eighty are pests of economic concern, including some of the world’s foremost fruit and vegetable pests, and there are many morphologically cryptic species complexes in the tribe. Where previous studies showed limited success in using COI to identify Dacini, our results with a highly curated morphological dataset indicate high congruence between morphology and COI: 90% of the species in our 5,576 sequences, 262-species global dataset can be identified with COI alone based on a monophyly criterion. However, in some key pest species belonging to complexes that were previously thought diagnosable with COI, we found that expanded sampling and independent validation of identifications using genomic data revealed introgression of mitochondrial DNA. We find that the informative SNPs are uniformly distributed across the COI gene, and we provide recommendations for standardization. We conclude that reliable molecular identifications with COI require extensive species coverage, population sampling, and genomics-supported reference identifications before they can be validated as a “diagnostic” marker for specific groups.