Tracking the origins of fly invasions; using mitochondrial haplotype diversity to identify potential source populations in two genetically intertwined fruit fly species (Bactrocera carambolae and Bactrocera dorsalis [Diptera: Tephritidae])
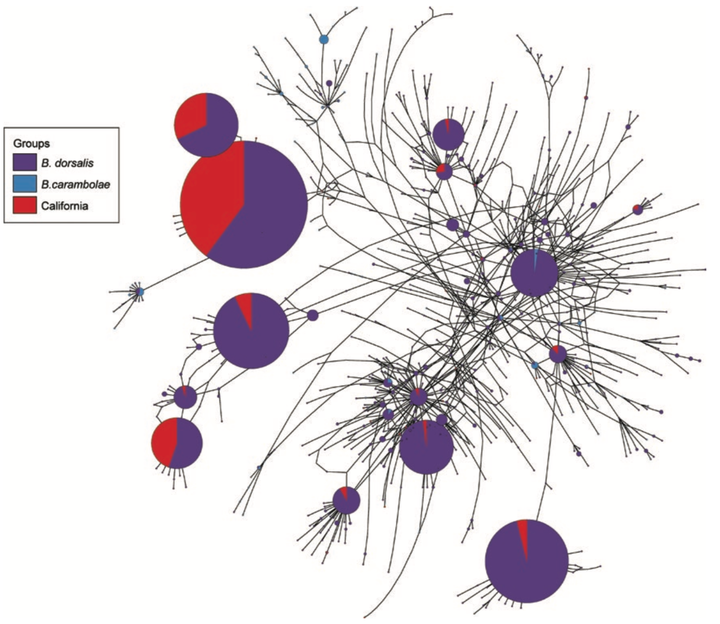 Image credit: M San Jose
Image credit: M San JoseAbstract
Bactrocera carambolae Drew and Hancock and Bactrocera dorsalis (Hendel) (Diptera: Tephritidae) are important pests of many fruits.These flies have been spread across the world through global travel and trade, and new areas are at risk of invasion. Whenever new invasive populations are discovered, quick and accurate identification is needed to mitigate the damage they can cause. Determining invasive pathways can prevent further spread of pests as well as subsequent reinvasions through the same pathway. Molecular markers can be used for both species identification and pathway analysis. We analyzed 1,601 individuals from 19 populations using 765 base pairs of the mitochondrial cytochrome oxidase I (COI) gene to infer the haplotype diversity and population structure within these flies from across their native and invasive ranges. We analyzed these samples by either grouping by species or geographic populations due to the genetic similarity in the mitochondrial genome. We found no genetic structure between B. dorsalis and B. carambolae and our findings suggest recent and most likely ongoing, genetic exchange between these two species in the wild. Hyper-diverse mitochondrial genetic diversity in the native range suggests large population sizes and relatively high mutation rates. Only 52% of the haplotypes found in the trap captures from California are shared with haplotypes from flies found in our global survey, indicating significant genetic diversity in the native range that is missing from our samples. However, these results provide a foundation for the accurate determination of the provenance of invasive populations around the world.