DNA barcoding of the leaf-mining moth subgenus Ectoedemia s. str. (Lepidoptera: Nepticulidae) with COI and EF1-alpha: two are better than one in recognising cryptic species
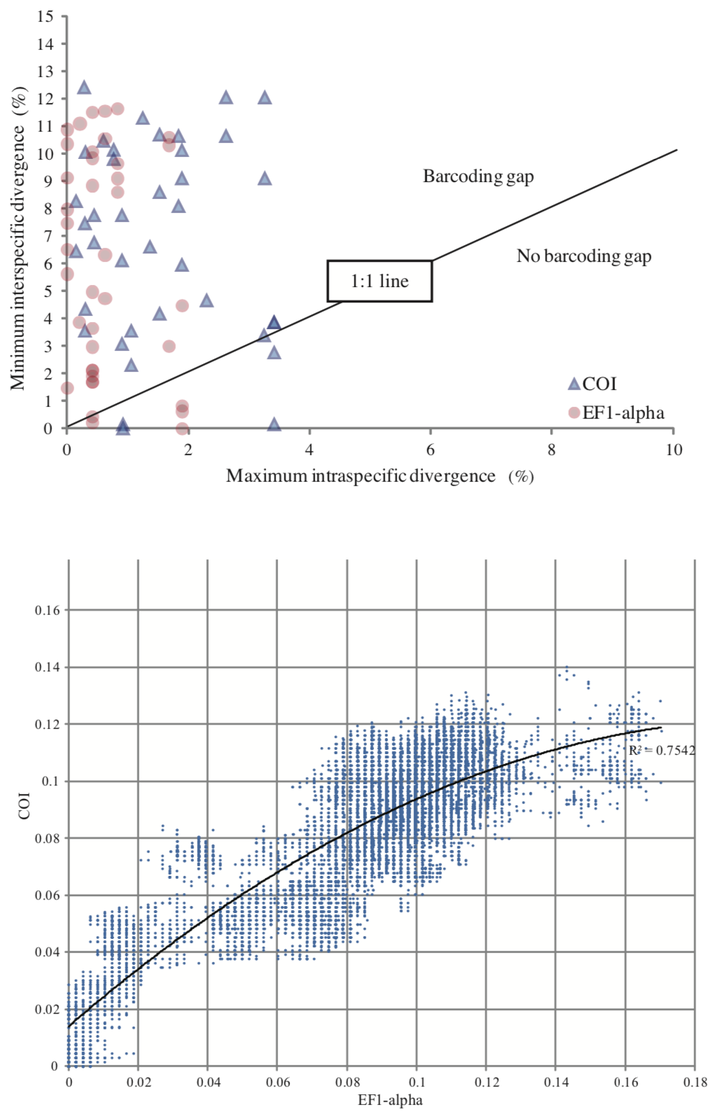 Image credit: C Doorenweerd
Image credit: C DoorenweerdAbstract
We sequenced 665bp of the Cytochrome C Oxidase I (COI) barcoding marker for 257 specimens and 482bp of Elongation Factor 1-α (EF1-α) for 237 specimens belonging to the leaf- mining subgenus Ectoedemia (Ectoedemia) in the basal Lepidopteran family Nepticulidae. The dataset includes 45 out of 48 West Palearctic Ectoedemia s. str. species and several species from Africa, North America and Asia. Both COI and EF1-α proved reliable as an alternative to conventional species identification for the majority of species and the combination of both markers can aid in species validation. A clear barcode gap is not present, and in some species large K2P intraspecific pairwise differences are found, up to 6.85% in COI and 2.9% in EF1-α. In the Ectoedemia rubivora species complex, the species E. rubivora, E. arcuatella and E. atricollis share COI barcodes and could only be distinguished by EF1-α. Diagnostic base positions, usually third codon positions, are in this and other cases a useful addition to species delimitation, in addition to distance methods. Ectoedemia albifasciella COI barcodes fall into two distinct clusters not related to other characters, whereas these clusters are absent in EF1-α, possibly caused by mtDNA anomalies or hybridisation. In the Ectoedemia subbimaculella complex, both sequences fail to unequivocally distinguish the species E. heringi, E. liechtensteini, E. phyllotomella and one population of E. subbimaculella. DNA barcodes confirm that North American Ectoedemia argyropeza are derived from a European introduction. We strongly advocate the use of a nuclear marker in addition to the universal COI barcode marker for better identifying species, including cryptic ones.